The title of this blog refers to the line in
the Beggar's Litany from around 1600, which I am fond of quoting to my many Yorkshire friends and
colleagues:
The title of this blog refers to the line in
the Beggar's Litany from around 1600, which I am fond of quoting to my many Yorkshire friends and
colleagues:
"From Hull, Hell and Halifax, may the Good Lord
deliver me"
The guillotine was in use regularly in Halifax over 400 years ago:
not just in Republican France! In fact it was used by the monarchy (well the
Royal Mint) to stamp out counterfeiting. When I have time I will tell you
about Sir Isaac Newton's other job for the Royal Mint! Getting back to
diseases....You will all be aware of the human tragedy playing out on the West
coast of Africa, where over a thousand people, young and
old have died following infection by the Ebola virus. The outbreak is all too
familiar to some of us who watched as the AIDS/HIV story played out in the
1980s
 And of course until very recently, Malaria held the inauspicious position as the world's biggest killer. The World Health Organisation (WHO) have only recently determined that Malaria has been overtaken by by cardiovascular disease (that's a topic for later!). Nevertheless, despite the many advances in modern medicine the human race remains highly vulnerable to parasitic diseases and viral infections. I constantly keep thinking that Achilles at least had only two heels that removed his invincibility in battle!
And of course until very recently, Malaria held the inauspicious position as the world's biggest killer. The World Health Organisation (WHO) have only recently determined that Malaria has been overtaken by by cardiovascular disease (that's a topic for later!). Nevertheless, despite the many advances in modern medicine the human race remains highly vulnerable to parasitic diseases and viral infections. I constantly keep thinking that Achilles at least had only two heels that removed his invincibility in battle!

Students returning and newcomers to the UTC
will be "welcomed" by an ambitious thematic programme that
incorporates experimental classes and enrichment in the Innovation Labs, aimed
at providing you with the opportunity to build your scientific skills both
technically and intellectually around some of the most challenging Health
related problems facing the world today. Before I give you a taster of what is
coming, let me say a few words about Ebola virus since it is in the news at the
moment.
 The picture on the left is one that the BBC are using a lot at the moment. It is a very high magnification image of ebola virus, which has been coloured artificially to enhance our understanding of its properties. Compared with the bacteriophage particles that we have come to know and love (humour me!), the shape is odd, it is more like an elongated bacillus, or rod-shaped bacterium, it is about 10% of the length of many mammalian cells, so it is pretty long. (the AIDS virus (below right, the green spiky sphere ) is quite different, almost spherical with a typical radius of 100nm, which is 10 times shorter than the length of ebola. A typical cell is around 10 000nm. Both viruses have an RNA genome, which means they have to trick the host cell into converting the genome into DNA first before they can begin to replicate, assemble and produce more infectious viral particles.
The picture on the left is one that the BBC are using a lot at the moment. It is a very high magnification image of ebola virus, which has been coloured artificially to enhance our understanding of its properties. Compared with the bacteriophage particles that we have come to know and love (humour me!), the shape is odd, it is more like an elongated bacillus, or rod-shaped bacterium, it is about 10% of the length of many mammalian cells, so it is pretty long. (the AIDS virus (below right, the green spiky sphere ) is quite different, almost spherical with a typical radius of 100nm, which is 10 times shorter than the length of ebola. A typical cell is around 10 000nm. Both viruses have an RNA genome, which means they have to trick the host cell into converting the genome into DNA first before they can begin to replicate, assemble and produce more infectious viral particles.

The ebola virus has only 7 genes, making it a very efficient beast! The AIDS virus has a few more. The closest relative of ebola is called Marburg virus, which is equally lethal. Why then can these viruses cause so much of a problem. There are two main reasons, one is Biological, but the second reason is just as important, if not more critical and that is Cultural practices. The virus attaches to cells found in the blood stream, fusing with the membrane and then executing a prolific programme of gene expression and replication, leading to an explosive infection that gives rise to a massive bleeding episode, from which recovery is challenging.
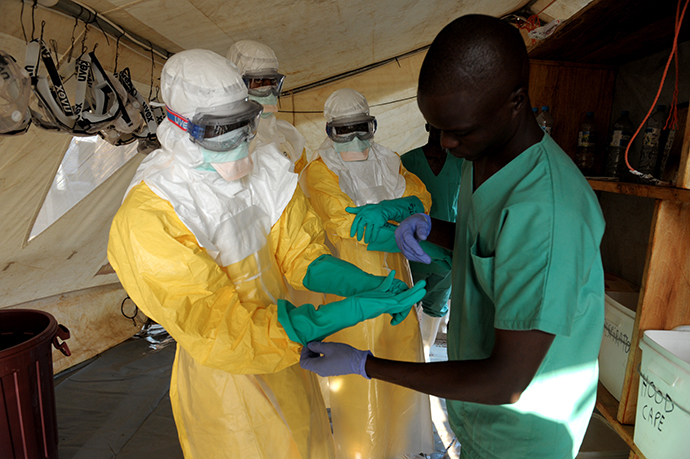 From a cultural perspective, the virus is spread through contaminated food: fruit bats are ebola resistant and therefore they spread the virus through fruit and droppings. Equally, infected animals eaten as "bush meat" are also sources of infection. Finally, families in the major hot spots in Africa have a culture in which sick members of the family are cared for at home, increasing the risk of spread. Poor education and even worse poor adherence to what we would consider normal hygiene routines, is probably at the heart of the spread of infection. The combination of the speed of pathology and the habits of the victims makes ebola, like AIDS before it a major challenge. You may be aware of the difficulty in persuading families to sleep under insecticide treated bed-nets, which would dramatically improve the prevention of malaria! So you must consider these diseases from both the Science and Social aspects.You must decide whether you think it is only appropriate to develop drugs and vaccines that are cost effective, typically for use in the West. In contrast, these potent diseases are prevalent in the poorest parts of the world, consequently resources are generally mobilised after a crisis! Compare that with the recent reaction to swine flu in the UK. If you are interested take a look at web sites like the Bill and Melinda Gates Foundation for more information.
From a cultural perspective, the virus is spread through contaminated food: fruit bats are ebola resistant and therefore they spread the virus through fruit and droppings. Equally, infected animals eaten as "bush meat" are also sources of infection. Finally, families in the major hot spots in Africa have a culture in which sick members of the family are cared for at home, increasing the risk of spread. Poor education and even worse poor adherence to what we would consider normal hygiene routines, is probably at the heart of the spread of infection. The combination of the speed of pathology and the habits of the victims makes ebola, like AIDS before it a major challenge. You may be aware of the difficulty in persuading families to sleep under insecticide treated bed-nets, which would dramatically improve the prevention of malaria! So you must consider these diseases from both the Science and Social aspects.You must decide whether you think it is only appropriate to develop drugs and vaccines that are cost effective, typically for use in the West. In contrast, these potent diseases are prevalent in the poorest parts of the world, consequently resources are generally mobilised after a crisis! Compare that with the recent reaction to swine flu in the UK. If you are interested take a look at web sites like the Bill and Melinda Gates Foundation for more information.
 The programme that awaits you at the UTC in September will give you an introduction to the Biology of parasites and infectious diseases, the socio-economic aspects and the opportunity to develop your laboratory skills by exploring model systems first hand. It is through these approaches that we have begun to understand the disease mechanisms, the interplay of their genomes and, importantly, to set some of you on the road to discovering a new generation of treatments for these virulent diseases. I am looking forward to helping you on this journey soon!
The programme that awaits you at the UTC in September will give you an introduction to the Biology of parasites and infectious diseases, the socio-economic aspects and the opportunity to develop your laboratory skills by exploring model systems first hand. It is through these approaches that we have begun to understand the disease mechanisms, the interplay of their genomes and, importantly, to set some of you on the road to discovering a new generation of treatments for these virulent diseases. I am looking forward to helping you on this journey soon!