So what's in the title? Well, we started the year looking a the differential growth of oral swabs on blood and nutrient agar plates (and blood forms the basis of Y11's innovation work in term 1 and part of the introduction to forensics for Y12s after the Easter vacation). Sweat and tears? Well, we use lysozyme to break down the bacterial cell wall (and you will recall that's where we secrete lysozyme, as a primitive anti-bacterial defence mechanism). And that leaves milk, which needs no further explanation!
So moving on to the Y12 project. Early discussions with senior members of the Eden Biodesign team, significantly enhanced the design of the Y12 project that centred on the controlled production and purification of a recombinant protein in a bacterial host. Eden (for short) produce recombinant polypeptides (another name for proteins) at level of purity far exceeding that achieved in our labs (since their products are often administered to patients!). We decided however, that the best initial approach was to introduce you to the fundamental principles of cell growth, gene regulation, selective gene induction, protein isolation and purification in a simple prokaryotic system first! Well, I can assure you that all of your final presentations and those selected for presentation to the Eden team, were really impressive.
In one sentence I have covered a significant part of an undergraduate degree course in Molecular Biology (an observation that was not unnoticed by the Eden team). So let's just take a few lines to explain the Science behind this project.
Escherichia coli were used as the bacterial host for the expression of a recombinant form of the Green Fluorescent protein, which is naturally expressed in certain jelly fish. One of my colleagues at the University of Sheffield, Professor Simon Foster passed on a bacterial strain harbouring a plasmid encoding this gene which is placed downstream of the lac operon. When you established that the optimum growth temperature was 37 degrees for the strain, you then induced expression of the GFP gene by "de-repressing" the lac promoter. Recall that the IPTG (a molecular mimic of the sugar lactose) prevents the lac repressor from blocking access to the host cell RNA Polymerase. As a consequence the gene for GFP is expressed, the protein is synthesised from its mRNA (on the ribosome) and by the end of week two, you all had a batch of cells in the freezer packed full of recombinant GFP.
So far so good. Time to move from Molecular Microbiology to Biochemistry (and since its my blog, the most important Life Science subject!). One of the tricks that have made Biochemistry so accessible (in particular protein biochemistry) to general molecular biologists, has been the application of genetics to add "tags", or additional sequences of amino acids that selectively bind to "ligands" (defined molecules ranging from metabolites to other proteins) that can be readily immobilised (attached) to the beads that make up column chromatography resins. We call this methodology affinity chromatography and we used the interaction between a poly Histidine tag (6 His codons were added to the GFP gene) and Nickel ions immobilised through a compound called NTA. This interaction is the basis of one of the most popular methods in protein purification in Biotechnology today.
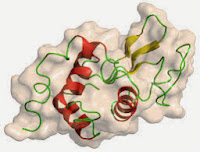
So, we have a cell pellet in the freezer, stuffed with GFP (His-tagged): the next step is to release the GFP (along with E.coli's own proteins that can be anything up to 2-3000 in number!). To do this we chose to add the enzyme lysozyme (obtained from egg white). It is possible to break cells by a range of chemical and mechanical methods, and you will use different methods in the Unilever project in January. Lysozyme is also an important landmark enzyme in the history of Biochemistry, since it was the first enzyme to have its 3D structure determined (above) by Sir David Phillips' group at Oxford in 1965.
The next step in the project was to use a column of Ni-NTA beads to capture the soluble recombinant GFP, while removing the contaminating host proteins. This was probably the point at which some of you went astray (although I estimate from the SDS PAGE experiment that more than 70% of the class obtained pure GFP). The difference was mainly the yield; which is commonly the case if you ask a new PhD student to make a batch of the lab's favourite recombinant protein, so nothing to worry about for a first attempt!
The tricky part of column chromatography is ensuring the resin stays fully immersed in buffer and that the sample loaded is not too dilute. In both ion exchange and affinity chromatography (unlike gel filtration chromatography), the material being purified is adsorbed to the resin and therefore becomes concentrated. This means you can get away with less attention to detail than when performing gel filtration chromatography, where the sample becomes increasingly dilute as it runs through the column. The elution step involved the displacement of the GFP bound to the Ni ions via the hexa-His tag. For this we used a high concentration (300mM) of imidazole (a mimic of His). The GFP was selectively displaced and samples collected in eppendorf tubes in 2-3 drop fractions. Again, this is challenging and I would say that around 10% of you obtained the GFP in a form that was sufficiently concentrated to see it glow in the lab. However as in the SDS PAGE gel at the left, most of the class produced an almost pure sample and I (and the Eden team) were duly impressed.
After you had all left Michael pooled the samples and we used a larger column to concentrate the "class GFP". As you can see below, one of my best Christmas presents was the bright, green preparation that was a fantastic achievement by you all!
So moving on to the Y12 project. Early discussions with senior members of the Eden Biodesign team, significantly enhanced the design of the Y12 project that centred on the controlled production and purification of a recombinant protein in a bacterial host. Eden (for short) produce recombinant polypeptides (another name for proteins) at level of purity far exceeding that achieved in our labs (since their products are often administered to patients!). We decided however, that the best initial approach was to introduce you to the fundamental principles of cell growth, gene regulation, selective gene induction, protein isolation and purification in a simple prokaryotic system first! Well, I can assure you that all of your final presentations and those selected for presentation to the Eden team, were really impressive.
In one sentence I have covered a significant part of an undergraduate degree course in Molecular Biology (an observation that was not unnoticed by the Eden team). So let's just take a few lines to explain the Science behind this project.
Escherichia coli were used as the bacterial host for the expression of a recombinant form of the Green Fluorescent protein, which is naturally expressed in certain jelly fish. One of my colleagues at the University of Sheffield, Professor Simon Foster passed on a bacterial strain harbouring a plasmid encoding this gene which is placed downstream of the lac operon. When you established that the optimum growth temperature was 37 degrees for the strain, you then induced expression of the GFP gene by "de-repressing" the lac promoter. Recall that the IPTG (a molecular mimic of the sugar lactose) prevents the lac repressor from blocking access to the host cell RNA Polymerase. As a consequence the gene for GFP is expressed, the protein is synthesised from its mRNA (on the ribosome) and by the end of week two, you all had a batch of cells in the freezer packed full of recombinant GFP.
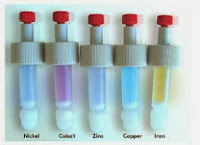 |
| A range of NTA columns containing different immobilised metal ions |
So, we have a cell pellet in the freezer, stuffed with GFP (His-tagged): the next step is to release the GFP (along with E.coli's own proteins that can be anything up to 2-3000 in number!). To do this we chose to add the enzyme lysozyme (obtained from egg white). It is possible to break cells by a range of chemical and mechanical methods, and you will use different methods in the Unilever project in January. Lysozyme is also an important landmark enzyme in the history of Biochemistry, since it was the first enzyme to have its 3D structure determined (above) by Sir David Phillips' group at Oxford in 1965.
The next step in the project was to use a column of Ni-NTA beads to capture the soluble recombinant GFP, while removing the contaminating host proteins. This was probably the point at which some of you went astray (although I estimate from the SDS PAGE experiment that more than 70% of the class obtained pure GFP). The difference was mainly the yield; which is commonly the case if you ask a new PhD student to make a batch of the lab's favourite recombinant protein, so nothing to worry about for a first attempt!
 |
| Several of the class preparations of Ni-NTA purified GFP. (Lane 6 in the middle is a marker) |
After you had all left Michael pooled the samples and we used a larger column to concentrate the "class GFP". As you can see below, one of my best Christmas presents was the bright, green preparation that was a fantastic achievement by you all!
 |
| A "class" act! |



No comments:
Post a Comment