 This
month I have been reading about sex determination in preparation for an
outreach project (of which more elsewhere). As a Biochemist, whenever I
want to learn something new in Biology, the
first thing that I do is search PubMed [subsection structure] for any
relevant molecules. In this case, the molecule that captured my interest
was the transcription factor encoded on the human Y chromosome which
had been shown around 25
years ago to be responsible for the determination of "maleness". The
protein in question is generally referred to as SRY, the abbreviated
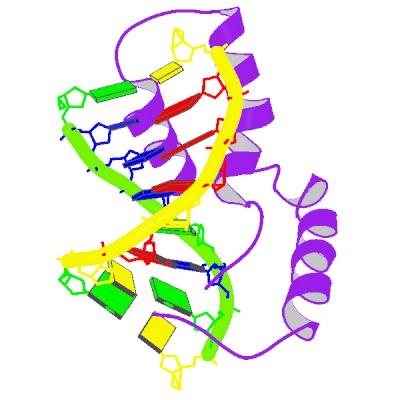
form of sex-determination region Y. The nice representation of SRY shown
top left, shows a purple polypeptide, interacting with a slightly
deformed double helical DNA sequence (shown in yellow and green). The
original structural work that I shall draw on comes from Michael Weiss's
laboratory originally at Harvard and now at Case Western in Ohio. together with work from Marius Clore's
group at the National Institutes for Health, in Maryland. The
structures were published quite some time ago and many insights have
been gained by comparative structural studies and also from subsequent
molecular, cellular, genetic and genomic work. In this short post, I
shall attempt to capture, the essence of the complexity of the role of
this otherwise rather simple, but nonetheless elegant molecule.
This
month I have been reading about sex determination in preparation for an
outreach project (of which more elsewhere). As a Biochemist, whenever I
want to learn something new in Biology, the
first thing that I do is search PubMed [subsection structure] for any
relevant molecules. In this case, the molecule that captured my interest
was the transcription factor encoded on the human Y chromosome which
had been shown around 25
years ago to be responsible for the determination of "maleness". The
protein in question is generally referred to as SRY, the abbreviated
form of sex-determination region Y. The nice representation of SRY shown
top left, shows a purple polypeptide, interacting with a slightly
deformed double helical DNA sequence (shown in yellow and green). The
original structural work that I shall draw on comes from Michael Weiss's
laboratory originally at Harvard and now at Case Western in Ohio. together with work from Marius Clore's
group at the National Institutes for Health, in Maryland. The
structures were published quite some time ago and many insights have
been gained by comparative structural studies and also from subsequent
molecular, cellular, genetic and genomic work. In this short post, I
shall attempt to capture, the essence of the complexity of the role of
this otherwise rather simple, but nonetheless elegant molecule.
 Let's begin with the biological problem. The first images of chromosomes were published in 1888 by
the German anatomist Heinrich Wilhelm Gottfried von Waldeyer-Hartz, who
also gave them their now familiar name. Around 20 years later, two
independent scientists Nettie Stevens (shown right) and Edmund Beecher
Wilson, demonstrated that the Y chromosome (as distinct from the X
chromosome) was the determinant of sex, in Nettie's case, she used the mealworm, Tenebrio molitor as her model system (which I have written about at length elsewhere).
It was Nettie who gave the chromosome the letter Y, not because it
looks a little like a Y, but because Y follows X, alphabetically. Roll
on around 90 years and the team of Robin Lovell Badge and Peter
Goodfellow at the National Institute for Medical Research in London,
demonstrated that the SRY gene on the Y chromosome was the testis
determining factor in humans. As a biological "footnote", the mechanisms
of sex determination (and gamete recognition) often show considerable
differences in otherwise quite similar species, making it challenging
(possibly futile?) to use model organisms in the study of reproductive
biology.
Think
for one minute what you would "expect" of a sex determination factor.
Given that the sequence (even before the structure was determined)
showed signs of it being a DNA binding protein, it makes sense for it to
be a "master regulator": a transcriptional regulator that perhaps, in
response to a series of hormonal "triggers", switches on (or off) the
expression of a group of genes that go on to establish the physiological
(and psychological?) features that define maleness. So, if it looks
like a DNA binding protein, does it bind to a specific nucleotide
sequence? And, perhaps such a sequence is found upstream of a set of
genes that are triggered, as discussed? These questions (and others)
have occupied those involved in unpacking the function of SRY over the
last 25 years.
Let's begin with the biological problem. The first images of chromosomes were published in 1888 by
the German anatomist Heinrich Wilhelm Gottfried von Waldeyer-Hartz, who
also gave them their now familiar name. Around 20 years later, two
independent scientists Nettie Stevens (shown right) and Edmund Beecher
Wilson, demonstrated that the Y chromosome (as distinct from the X
chromosome) was the determinant of sex, in Nettie's case, she used the mealworm, Tenebrio molitor as her model system (which I have written about at length elsewhere).
It was Nettie who gave the chromosome the letter Y, not because it
looks a little like a Y, but because Y follows X, alphabetically. Roll
on around 90 years and the team of Robin Lovell Badge and Peter
Goodfellow at the National Institute for Medical Research in London,
demonstrated that the SRY gene on the Y chromosome was the testis
determining factor in humans. As a biological "footnote", the mechanisms
of sex determination (and gamete recognition) often show considerable
differences in otherwise quite similar species, making it challenging
(possibly futile?) to use model organisms in the study of reproductive
biology.
Think
for one minute what you would "expect" of a sex determination factor.
Given that the sequence (even before the structure was determined)
showed signs of it being a DNA binding protein, it makes sense for it to
be a "master regulator": a transcriptional regulator that perhaps, in
response to a series of hormonal "triggers", switches on (or off) the
expression of a group of genes that go on to establish the physiological
(and psychological?) features that define maleness. So, if it looks
like a DNA binding protein, does it bind to a specific nucleotide
sequence? And, perhaps such a sequence is found upstream of a set of
genes that are triggered, as discussed? These questions (and others)
have occupied those involved in unpacking the function of SRY over the
last 25 years.
 The
SRY protein belongs to a class of non-histone, DNA binding proteins
called (for historical reasons) High Mobility Group Proteins. Very
simply, these proteins were first identified as "fast migrating"
non-histone species (see the species at the foot of the SDS gel on the
left), when purified by ion exchange chromatography and analysed by gel electrophoresis (initially by Goodwin and Johns,
in 1973). Today, the name is abbreviated (unhelpfully for newcomers to
the field!) to HMG or HMG Box proteins: several pioneering NMR groups
focused on the HMG Box proteins for their structural work, partly
because of their relatively small size and high solubility.I have spent
many an evening in Portsmouth talking about HMG proteins with Professor
Colin Crane-Robinson and his colleagues in the excellent Biophysics
group!
The
SRY protein belongs to a class of non-histone, DNA binding proteins
called (for historical reasons) High Mobility Group Proteins. Very
simply, these proteins were first identified as "fast migrating"
non-histone species (see the species at the foot of the SDS gel on the
left), when purified by ion exchange chromatography and analysed by gel electrophoresis (initially by Goodwin and Johns,
in 1973). Today, the name is abbreviated (unhelpfully for newcomers to
the field!) to HMG or HMG Box proteins: several pioneering NMR groups
focused on the HMG Box proteins for their structural work, partly
because of their relatively small size and high solubility.I have spent
many an evening in Portsmouth talking about HMG proteins with Professor
Colin Crane-Robinson and his colleagues in the excellent Biophysics
group!
 I have relied heavily on the excellent review on SRY by Kashimada and Koopman for the "Biology" in this post. Briefly, SRY acts to up-regulate expression of the gene called Sox9 (SRY Box [containing, gene] 9). The human SRY specifically binds to the nucleotide sequence (A/T)ACAA(T/A) in the minor groove of DNA, inducing a 60-85° bend. It has been shown that specific recognition AND induced bending of the DNA are essential for function (Sox9 activation). What follows in the Sertoli cell precursors is a cascade of events which stimulates expression of several other genes involved
in the differentiation of Sertoli cells (the diagram above captures the events, but the details can be found by a simple Google search, for the physiologists among you). It seems that SRY activity also suppresses the female sex-determining pathway (it is unclear to me whether this is an indirect or direct effect). Sertoli cells then stimulate the ultimate formation of the testes: the primary manifestation of maleness (I think?).
As I said earlier, the genes and proteins associated with reproductive biology represent a challenge for developmental biologists, who have relied heavily on the use of model organisms
to explore evolutionary aspects of such phenomena as limb formation,
brain development etc. Fortunately, although there are differences
between the murine and human SRY genes, there
are sufficient similarities (especially with respect to the HMG-Box
domain) that give us confidence that the major features of the testis determining pathways are conserved. So let's get back to this DNA recognition and "bending" phenomena.
I have relied heavily on the excellent review on SRY by Kashimada and Koopman for the "Biology" in this post. Briefly, SRY acts to up-regulate expression of the gene called Sox9 (SRY Box [containing, gene] 9). The human SRY specifically binds to the nucleotide sequence (A/T)ACAA(T/A) in the minor groove of DNA, inducing a 60-85° bend. It has been shown that specific recognition AND induced bending of the DNA are essential for function (Sox9 activation). What follows in the Sertoli cell precursors is a cascade of events which stimulates expression of several other genes involved
in the differentiation of Sertoli cells (the diagram above captures the events, but the details can be found by a simple Google search, for the physiologists among you). It seems that SRY activity also suppresses the female sex-determining pathway (it is unclear to me whether this is an indirect or direct effect). Sertoli cells then stimulate the ultimate formation of the testes: the primary manifestation of maleness (I think?).
As I said earlier, the genes and proteins associated with reproductive biology represent a challenge for developmental biologists, who have relied heavily on the use of model organisms
to explore evolutionary aspects of such phenomena as limb formation,
brain development etc. Fortunately, although there are differences
between the murine and human SRY genes, there
are sufficient similarities (especially with respect to the HMG-Box
domain) that give us confidence that the major features of the testis determining pathways are conserved. So let's get back to this DNA recognition and "bending" phenomena.
 Early structural studies on DNA binding proteins suggested that a two helix motif (the helix-turn-helix domain) provided an elegant solution to the sequence
-specific recognition of DNA (left). However, as is the normal course
of events in Biology, more structural studies revealed that there are a
variety of ways that DNA can be recognised; moreover, recognition does
not always leave Watson and Crick intact! In fact the recognition of DNA in eukaryotes by the histone proteins cannot leave the DNA structure unaffected, otherwise it would look like a long, insulated cable, rather than the compacted, space-saving arrangement we call chromatin. However, histones are for another post. The HMG-Box binding proteins like SRY induce a distortion in the DNA, but why? And just how difficult is it to "unsettle" what we have come to know and love as one of the most satisfying arrangements of the two polynucleotide chains.
Early structural studies on DNA binding proteins suggested that a two helix motif (the helix-turn-helix domain) provided an elegant solution to the sequence
-specific recognition of DNA (left). However, as is the normal course
of events in Biology, more structural studies revealed that there are a
variety of ways that DNA can be recognised; moreover, recognition does
not always leave Watson and Crick intact! In fact the recognition of DNA in eukaryotes by the histone proteins cannot leave the DNA structure unaffected, otherwise it would look like a long, insulated cable, rather than the compacted, space-saving arrangement we call chromatin. However, histones are for another post. The HMG-Box binding proteins like SRY induce a distortion in the DNA, but why? And just how difficult is it to "unsettle" what we have come to know and love as one of the most satisfying arrangements of the two polynucleotide chains.
 Watson and Crick's interpretation of the X-ray data of Franklin and others, the base composition analysis of Chargaff, the weight of circumstantial chemical and biological evidence and the emerging realisation that helices could underpin protein structures, all contributed to the appeal of the double helix. The follow up biological and biophysical experiments that set out to provide a strong foundation for the primacy of the double helix in molecular genetics, have largely confirmed Watson and Crick's ideas. However, as Watson and Crick themselves pointed out, the double helix must be unwound, however transiently, to facilitate DNA replication. Through the combination of creative experimental work by Biochemists, Biophysicists, Chemists, Geneticists and Cell Biologists, in conjunction with crystallographers and NMR spectroscopists, we now know that the iconic double helix is only part of the story. You can read here about one example of how the structure of DNA is very discretely perturbed in order to accommodate the addition of one of the epigenetic marks: methylated cytosine. Here we shall take a little time to understand DNA kinking. I have chosen to avoid the word bending, since, rightly or wrongly, I imagine bending to result from application of forces at two points flanking the bend. Imagine a 1m bamboo cane (above right), it is easy to bend by grabbing both ends and pushing towards the centre. It is much harder to bend if you hold
the cane at the centre with two hands! However, many DNA binding
proteins seem to induce kinking at the centre of that kinking.
Watson and Crick's interpretation of the X-ray data of Franklin and others, the base composition analysis of Chargaff, the weight of circumstantial chemical and biological evidence and the emerging realisation that helices could underpin protein structures, all contributed to the appeal of the double helix. The follow up biological and biophysical experiments that set out to provide a strong foundation for the primacy of the double helix in molecular genetics, have largely confirmed Watson and Crick's ideas. However, as Watson and Crick themselves pointed out, the double helix must be unwound, however transiently, to facilitate DNA replication. Through the combination of creative experimental work by Biochemists, Biophysicists, Chemists, Geneticists and Cell Biologists, in conjunction with crystallographers and NMR spectroscopists, we now know that the iconic double helix is only part of the story. You can read here about one example of how the structure of DNA is very discretely perturbed in order to accommodate the addition of one of the epigenetic marks: methylated cytosine. Here we shall take a little time to understand DNA kinking. I have chosen to avoid the word bending, since, rightly or wrongly, I imagine bending to result from application of forces at two points flanking the bend. Imagine a 1m bamboo cane (above right), it is easy to bend by grabbing both ends and pushing towards the centre. It is much harder to bend if you hold
the cane at the centre with two hands! However, many DNA binding
proteins seem to induce kinking at the centre of that kinking.
 The first examples of protein induced, DNA kinking that I came across were in the field of DNA restriction and modification. The restriction enzymes EcoRI and EcoRV, finally yielded to the crystallographers when synthetic DNA became widely available in the 1980s-'90s. Both of these enzymes hydrolyse DNA, with the help of Mg ions. The DNA adopts a kinked structure midway through the reaction pathway.
There are significant differences in the conformation of the kinked DNA
in both cases, but I think it is easy to get the logic of an enzyme
applying "pressure" at the break point. The image on the left shows the
outcome of Sac7d interacting with DNA, through the minor groove. It shows the striking kink really well. The promoter binding factor (TATA binding protein, or TBP is another great example of a promoter of kinks). Now just think about the consequences of kinking. Just bend your knee. The
skin on your knee cap gets stretched and the space behind gets
squeezed. The nice equilibrium has been upset. [Not forgetting that you
need to expend energy to bend your knee]. When molecules are in solution
in their lowest free energy state, it takes energy to disturb that
equilibrium, and a price must be paid. In Biological systems where we
can't always use heat exchange to solve our thermodynamic problems, we
often resort to an entropic solution. Ordered water molecules becomes
disordered as a consequence of a binding interaction, hence there is a
net increase in entropy [enough to stabilise the kinked complex, in this case]. We rationalise such distortions,
on the assumption that the complex structure is more stable than the two
components (the
protein and the B-form DNA) alone. It is important to remember that in
solution most equilibria are "dynamic equilibria", so that the observed
structure is the most common, or likely to populate the solution
at any one time. However, B form DNA is believed to make "excursions"
into other forms, which may well be selectively identified and captured by a DNA binding protein.
The first examples of protein induced, DNA kinking that I came across were in the field of DNA restriction and modification. The restriction enzymes EcoRI and EcoRV, finally yielded to the crystallographers when synthetic DNA became widely available in the 1980s-'90s. Both of these enzymes hydrolyse DNA, with the help of Mg ions. The DNA adopts a kinked structure midway through the reaction pathway.
There are significant differences in the conformation of the kinked DNA
in both cases, but I think it is easy to get the logic of an enzyme
applying "pressure" at the break point. The image on the left shows the
outcome of Sac7d interacting with DNA, through the minor groove. It shows the striking kink really well. The promoter binding factor (TATA binding protein, or TBP is another great example of a promoter of kinks). Now just think about the consequences of kinking. Just bend your knee. The
skin on your knee cap gets stretched and the space behind gets
squeezed. The nice equilibrium has been upset. [Not forgetting that you
need to expend energy to bend your knee]. When molecules are in solution
in their lowest free energy state, it takes energy to disturb that
equilibrium, and a price must be paid. In Biological systems where we
can't always use heat exchange to solve our thermodynamic problems, we
often resort to an entropic solution. Ordered water molecules becomes
disordered as a consequence of a binding interaction, hence there is a
net increase in entropy [enough to stabilise the kinked complex, in this case]. We rationalise such distortions,
on the assumption that the complex structure is more stable than the two
components (the
protein and the B-form DNA) alone. It is important to remember that in
solution most equilibria are "dynamic equilibria", so that the observed
structure is the most common, or likely to populate the solution
at any one time. However, B form DNA is believed to make "excursions"
into other forms, which may well be selectively identified and captured by a DNA binding protein.
 The HMG-Box proteins, including SRY, induce a kink in the target site via a three helix recognition motif (top figure) and in doing so establish conditions for the transcriptional apparatus, including RNA Polymerase II, to initiate transcription form the Sox9 gene. How do we know that DNA distortion is essential for the action
of SRY? There are mutations that allow the SRY protein to bind, but not
to bend (sorry, kinking seemed less appropriate!). You can read about
the detailed analysis of SRY mutants here. However it seems that as the data from genome analysis are pointing to pleiotropic effects of many mutations that we considered as explicable by localised defects in function. It is certainly the case that individual amino acids when substituted by mutation can have an impact on the recognition events, but such mutations may also make the protein more prone to degradation, or may interfere with other protein:protein
interactions for example. Nevertheless, the SRY DNA binding domain
provides an example of a simple solution to initiating one of the most striking phenomena in human biology: sex determination.
Finally, with respect to the title of my post, do I think that SRY is responsible for sex determination? Of course, the presence of a functional SRY is required to initiate the development of the male genitalia. Is sexuality black and white? Just as we are realising that point mutations may be explained at one level by local physicochemical effects, systems biology and genomics are uncovering contextual effects that suggest to me that sex is more likely to be less black and white and more "Fifty Shades of Grey"
The HMG-Box proteins, including SRY, induce a kink in the target site via a three helix recognition motif (top figure) and in doing so establish conditions for the transcriptional apparatus, including RNA Polymerase II, to initiate transcription form the Sox9 gene. How do we know that DNA distortion is essential for the action
of SRY? There are mutations that allow the SRY protein to bind, but not
to bend (sorry, kinking seemed less appropriate!). You can read about
the detailed analysis of SRY mutants here. However it seems that as the data from genome analysis are pointing to pleiotropic effects of many mutations that we considered as explicable by localised defects in function. It is certainly the case that individual amino acids when substituted by mutation can have an impact on the recognition events, but such mutations may also make the protein more prone to degradation, or may interfere with other protein:protein
interactions for example. Nevertheless, the SRY DNA binding domain
provides an example of a simple solution to initiating one of the most striking phenomena in human biology: sex determination.
Finally, with respect to the title of my post, do I think that SRY is responsible for sex determination? Of course, the presence of a functional SRY is required to initiate the development of the male genitalia. Is sexuality black and white? Just as we are realising that point mutations may be explained at one level by local physicochemical effects, systems biology and genomics are uncovering contextual effects that suggest to me that sex is more likely to be less black and white and more "Fifty Shades of Grey"
 This
month I have been reading about sex determination in preparation for an
outreach project (of which more elsewhere). As a Biochemist, whenever I
want to learn something new in Biology, the
first thing that I do is search PubMed [subsection structure] for any
relevant molecules. In this case, the molecule that captured my interest
was the transcription factor encoded on the human Y chromosome which
had been shown around 25
years ago to be responsible for the determination of "maleness". The
protein in question is generally referred to as SRY, the abbreviated
form of sex-determination region Y. The nice representation of SRY shown
top left, shows a purple polypeptide, interacting with a slightly
deformed double helical DNA sequence (shown in yellow and green). The
original structural work that I shall draw on comes from Michael Weiss's
laboratory originally at Harvard and now at Case Western in Ohio. together with work from Marius Clore's
group at the National Institutes for Health, in Maryland. The
structures were published quite some time ago and many insights have
been gained by comparative structural studies and also from subsequent
molecular, cellular, genetic and genomic work. In this short post, I
shall attempt to capture, the essence of the complexity of the role of
this otherwise rather simple, but nonetheless elegant molecule.
This
month I have been reading about sex determination in preparation for an
outreach project (of which more elsewhere). As a Biochemist, whenever I
want to learn something new in Biology, the
first thing that I do is search PubMed [subsection structure] for any
relevant molecules. In this case, the molecule that captured my interest
was the transcription factor encoded on the human Y chromosome which
had been shown around 25
years ago to be responsible for the determination of "maleness". The
protein in question is generally referred to as SRY, the abbreviated
form of sex-determination region Y. The nice representation of SRY shown
top left, shows a purple polypeptide, interacting with a slightly
deformed double helical DNA sequence (shown in yellow and green). The
original structural work that I shall draw on comes from Michael Weiss's
laboratory originally at Harvard and now at Case Western in Ohio. together with work from Marius Clore's
group at the National Institutes for Health, in Maryland. The
structures were published quite some time ago and many insights have
been gained by comparative structural studies and also from subsequent
molecular, cellular, genetic and genomic work. In this short post, I
shall attempt to capture, the essence of the complexity of the role of
this otherwise rather simple, but nonetheless elegant molecule.  Let's begin with the biological problem. The first images of chromosomes were published in 1888 by
the German anatomist Heinrich Wilhelm Gottfried von Waldeyer-Hartz, who
also gave them their now familiar name. Around 20 years later, two
independent scientists Nettie Stevens (shown right) and Edmund Beecher
Wilson, demonstrated that the Y chromosome (as distinct from the X
chromosome) was the determinant of sex, in Nettie's case, she used the mealworm, Tenebrio molitor as her model system (which I have written about at length elsewhere).
It was Nettie who gave the chromosome the letter Y, not because it
looks a little like a Y, but because Y follows X, alphabetically. Roll
on around 90 years and the team of Robin Lovell Badge and Peter
Goodfellow at the National Institute for Medical Research in London,
demonstrated that the SRY gene on the Y chromosome was the testis
determining factor in humans. As a biological "footnote", the mechanisms
of sex determination (and gamete recognition) often show considerable
differences in otherwise quite similar species, making it challenging
(possibly futile?) to use model organisms in the study of reproductive
biology.
Let's begin with the biological problem. The first images of chromosomes were published in 1888 by
the German anatomist Heinrich Wilhelm Gottfried von Waldeyer-Hartz, who
also gave them their now familiar name. Around 20 years later, two
independent scientists Nettie Stevens (shown right) and Edmund Beecher
Wilson, demonstrated that the Y chromosome (as distinct from the X
chromosome) was the determinant of sex, in Nettie's case, she used the mealworm, Tenebrio molitor as her model system (which I have written about at length elsewhere).
It was Nettie who gave the chromosome the letter Y, not because it
looks a little like a Y, but because Y follows X, alphabetically. Roll
on around 90 years and the team of Robin Lovell Badge and Peter
Goodfellow at the National Institute for Medical Research in London,
demonstrated that the SRY gene on the Y chromosome was the testis
determining factor in humans. As a biological "footnote", the mechanisms
of sex determination (and gamete recognition) often show considerable
differences in otherwise quite similar species, making it challenging
(possibly futile?) to use model organisms in the study of reproductive
biology.