 As usual, I had a molecule almost ready to go, until I started reading an interesting thesis from one of (Professor) Mike McPherson's research group at the University of Leeds. Without disclosing any secrets, the thesis centred around a class of molecules called adhirons (see the left hand image from the Leeds group RCSB PDB). I thought I would put my reading to good use and share my enthusiasm for these remarkable polypeptides. On the one hand they offer an excellent opportunity to challenge the primacy of antibodies in high affinity, selective protein binding, but they also offer considerable insight into the evolutionary relationships between primary structure and the "encoded" tertiary interactions between protein surfaces. And as you know, it is the network of protein:protein interactions that expands the functionality of many eukaryotic genomes. Since "Molecule of the Month" aims to introduce you to a mixture of "Old Chestnuts" and "Young Turks", I also try to select molecules for the lessons they can teach about the relationship between protein structure and function in an evolutionary context.
As usual, I had a molecule almost ready to go, until I started reading an interesting thesis from one of (Professor) Mike McPherson's research group at the University of Leeds. Without disclosing any secrets, the thesis centred around a class of molecules called adhirons (see the left hand image from the Leeds group RCSB PDB). I thought I would put my reading to good use and share my enthusiasm for these remarkable polypeptides. On the one hand they offer an excellent opportunity to challenge the primacy of antibodies in high affinity, selective protein binding, but they also offer considerable insight into the evolutionary relationships between primary structure and the "encoded" tertiary interactions between protein surfaces. And as you know, it is the network of protein:protein interactions that expands the functionality of many eukaryotic genomes. Since "Molecule of the Month" aims to introduce you to a mixture of "Old Chestnuts" and "Young Turks", I also try to select molecules for the lessons they can teach about the relationship between protein structure and function in an evolutionary context.Adhirons and antibodies have some things in common and some significant differences. I have written about IgG before, and rather than give details, I shall cite their general properties for comparative purposes here, since antibodies form a major part of any Biology curriculum in High School and above. The first point to make is that Adhirons are not found in Nature. However, they are molecules that have been considerably "informed" by Nature. They are derivatives of a group of protease inhibitors referred to as statins (but not the cardiovascular drugs), a class of cysteine protease inhibitors which are related to the stefins, studied in Sheffield in (Professor) Jon Waltho's structural biology group in the 1990s. The development of NMR for the determination of protein structure has been helped significantly by the existence in Nature of polypeptides of less than 100 amino acids (low molecular weight) that exist to keep hydrolytic enzymes, including proteases, nucleases and amylases in check. Such proteins (Tendamistat, Bovine Pancreatic Trypsin Inhibitor and Statins etc.) proved to be workhorses for method development and helped interpretation of the complex spectra derived from sophisticated NMR experiments. You can read Nobel laureate, Kurt Wüthrich's overview of the development of NMR in structural biology here.
The adhiron used in Mike's group is derived from a plant statin family called the phytostatins. It was selected for its simplicity (no awkward post-translational modifications), and its intrinsic physical properties. It is stable at elevated temperatures (around 100 degrees C), it is extremely soluble (hundred of mg/ml solutions are not uncommon: many proteins drop out of solution above concentrations of 1mg/ml!), it has no sensitive disulphide bonds (which can lead to redox problems in use) and it is relatively low in molecular weight and monomeric, which means every application becomes simplified and that much more cost-effective. You can read more about the background to the development of adhirons here.
It is less intuitive, to appreciate how two flexible surfaces can unite and organise rigidly (which is what enzyme active sites have to do most of the time). But think of a mixture of oil and water: although the two liquids are "mobile"; the interface is very sharp. Or think of pushing your hand into a soft glove. The other way I think of this challenging phenomenon is to consider protein folding during translation. As a polypeptide chain emerges from the ribosome it must find and adopt its tertiary structure pretty quickly (life and death is less than 30 minutes in E.coli!). So a disordered region, meeting a ligand or another protein surface will harness the forces of entropy and enthalpy in the same way as the folding polypeptdide. The role of the scaffold in antibodies and adhirons, serves to facilitate this: and experimental observations have shown that this strategy is really successful in other settings, including RNA binding and sequence-specific DNA recognition. It is this approach to molecular recognition that is adopted by adhirons. And this is translated into dissociation constants between adhirons and their targets that are often in the tens of picomolar range. [As a rule of thumb, you can estimate a dissociation constant (recall lower the molarity, the higher the affinity) from the average physiological concentration of the ligand: substrates for enzymes have Kds of a few mM, coenzymes are found at tens of micromolar and individual proteins and nucleic acids have Kds in the nM range)].
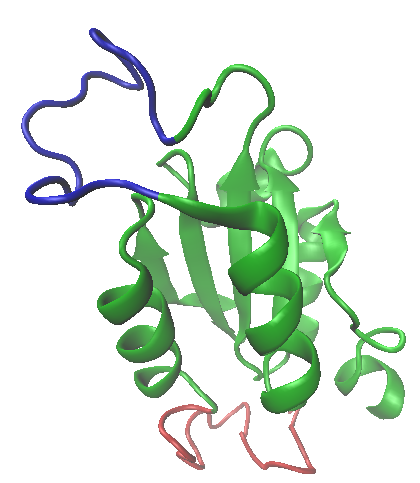 The variation in antibody specificity arises owing to the expression of a diverse set of variable regions that are linked to the constant domain (I wont discuss the mechanism here, but it is something you should read about). Adhirons can be generated in vitro that will specifically bind to many biological targets (although I expect not all with the same affinity), by randomising the loop regions that form the "spanner in the works" as statins block cysteine proteases. This can be achieved experimentally using an error prone DNA polymerase or some form of synthetic DNA, localised gene replacement method. The "library" of mutants can then be used to screen the target: a favourite technique for this is known as phage display, and you can read the details here. In this way, high affinity, highly selective adhirons can be generated to recognise target molecules with all of the advantages of a monoclonal antibody, but at a much lower cost (in view of the relatively simple methodology). The combination of a flexible interaction surface supported by a stable and relatively invariant scaffold domain in proteins, represents an interesting evolutionary concept that Nature has exploited on several occasions. (One such scaffold and loop structure is shown top left, which is part of the structure of thioredoxin, a widely distributed redox protein: the loop regions is shown in blue). Adhirons are not in the mainstream of molecular recognition reagents just yet, and the technologies for detection etc. that have grown up around antibodies are certainly not about to become obsolete, but these molecules certainly seem to me to offer some very significant opportunities for research and diagnostics in the future.
The variation in antibody specificity arises owing to the expression of a diverse set of variable regions that are linked to the constant domain (I wont discuss the mechanism here, but it is something you should read about). Adhirons can be generated in vitro that will specifically bind to many biological targets (although I expect not all with the same affinity), by randomising the loop regions that form the "spanner in the works" as statins block cysteine proteases. This can be achieved experimentally using an error prone DNA polymerase or some form of synthetic DNA, localised gene replacement method. The "library" of mutants can then be used to screen the target: a favourite technique for this is known as phage display, and you can read the details here. In this way, high affinity, highly selective adhirons can be generated to recognise target molecules with all of the advantages of a monoclonal antibody, but at a much lower cost (in view of the relatively simple methodology). The combination of a flexible interaction surface supported by a stable and relatively invariant scaffold domain in proteins, represents an interesting evolutionary concept that Nature has exploited on several occasions. (One such scaffold and loop structure is shown top left, which is part of the structure of thioredoxin, a widely distributed redox protein: the loop regions is shown in blue). Adhirons are not in the mainstream of molecular recognition reagents just yet, and the technologies for detection etc. that have grown up around antibodies are certainly not about to become obsolete, but these molecules certainly seem to me to offer some very significant opportunities for research and diagnostics in the future.Summary of Key Points
No comments:
Post a Comment