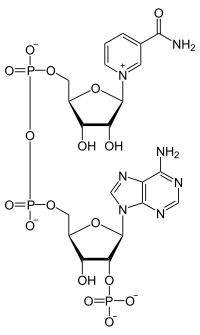
 Biosynthesis of NAD. Two British Biochemists(Harden and Young), working at the newly formed Lister Institute in the first part of the last century, discovered a heat stable molecule that stimulated a number of yeast fermentation reactions. They called this extract a coferment. We now know that this coenzyme is synthesized in mammals, plants and bacteria, but in slightly different ways. Indeed these often complementary aspects of coenzyme biosynthesis provide one of the many reasons why our own microbiota prove beneficial. In all organisms, amino acids generally provide the starting materials for the biosynthesis of molecules like NAD. In man, the starting point is the two ringed amino acid, tryptophan (Trp or W): in bacteria the acidic amino acid aspartate (Asp or D) may be the starting point. However the first key step is the production of the dicarboxylate, quinolinic acid: a pyridine ring, with two carboxylic acid substituents at 2 o'clock and 4 o'clock (technically pyridine 2,3 dicarboxylic acid). The structure of quinolinic acid is shown top left, and you should note the Nitrogen, which is a key atom in this molecule (and subsequently in NAD). Following the addition of a ribose phosphate and an adenylate, the final step on the way to NAD is the amidation of the pyridine ring. To make NADP, the enzyme NAD kinase obliges. Finally, I should mention the concept of salvage pathways, which has become something of a biochemical backwater, since I was a student. However this is a simple concept: if you eat food that contains fragments of NAD (or indeed molecules like ATP or CTP etc), you can top up your NAD levels through a group of so called salvage pathways. A nice evolutionary bonus!
Biosynthesis of NAD. Two British Biochemists(Harden and Young), working at the newly formed Lister Institute in the first part of the last century, discovered a heat stable molecule that stimulated a number of yeast fermentation reactions. They called this extract a coferment. We now know that this coenzyme is synthesized in mammals, plants and bacteria, but in slightly different ways. Indeed these often complementary aspects of coenzyme biosynthesis provide one of the many reasons why our own microbiota prove beneficial. In all organisms, amino acids generally provide the starting materials for the biosynthesis of molecules like NAD. In man, the starting point is the two ringed amino acid, tryptophan (Trp or W): in bacteria the acidic amino acid aspartate (Asp or D) may be the starting point. However the first key step is the production of the dicarboxylate, quinolinic acid: a pyridine ring, with two carboxylic acid substituents at 2 o'clock and 4 o'clock (technically pyridine 2,3 dicarboxylic acid). The structure of quinolinic acid is shown top left, and you should note the Nitrogen, which is a key atom in this molecule (and subsequently in NAD). Following the addition of a ribose phosphate and an adenylate, the final step on the way to NAD is the amidation of the pyridine ring. To make NADP, the enzyme NAD kinase obliges. Finally, I should mention the concept of salvage pathways, which has become something of a biochemical backwater, since I was a student. However this is a simple concept: if you eat food that contains fragments of NAD (or indeed molecules like ATP or CTP etc), you can top up your NAD levels through a group of so called salvage pathways. A nice evolutionary bonus! Having manufactured NAD either de novo, or via a salvage process, the coenzyme participates in numerous enzyme catalysed reactions. I have already mentioned the GDH reaction, but in my recent post about metabolism, it was clear to the metabolic pioneers at the turn of the last century, that a 2 H transfer reaction was likely to be a common feature in the conversion of the food we eat into energy. NAD binds tightly (with dissociation constants in the sub-millimolar range) to dehygrogenase enzymes that recognise alcohols, glyceraldehyd-3-phosphate, sugars and fatty acids to name a few molecules that have the functional term "dehydrogenase" appended. (I should note that NADH generally binds more tightly than NAD). The binding site for NAD is often called the Rossman fold (see the image above on the RHS: the red molecule is NAD), after the eminent crystallographer, Michael Rossman, who studied these classes of enzymes in the 1970s. The mechanism of dhydrogenases differs subtly between the different "classes", but all share a common feature: the positively charged Nitrogen accepts a hydride from the substrate. In some enzymes the hydride is transferred above (A) and in others below (B) the plane of the pyridine ring. The other proton is usually bound to an amino acid side chain. The reverse is true for the NADH to NAD direction. If you are interested in the details of dehydrogenase enzymes, there are many excellent sections in text books, or reviews: I like the descriptions in Alan Fersht's classic text and this web site will give you more detailed information.
Having manufactured NAD either de novo, or via a salvage process, the coenzyme participates in numerous enzyme catalysed reactions. I have already mentioned the GDH reaction, but in my recent post about metabolism, it was clear to the metabolic pioneers at the turn of the last century, that a 2 H transfer reaction was likely to be a common feature in the conversion of the food we eat into energy. NAD binds tightly (with dissociation constants in the sub-millimolar range) to dehygrogenase enzymes that recognise alcohols, glyceraldehyd-3-phosphate, sugars and fatty acids to name a few molecules that have the functional term "dehydrogenase" appended. (I should note that NADH generally binds more tightly than NAD). The binding site for NAD is often called the Rossman fold (see the image above on the RHS: the red molecule is NAD), after the eminent crystallographer, Michael Rossman, who studied these classes of enzymes in the 1970s. The mechanism of dhydrogenases differs subtly between the different "classes", but all share a common feature: the positively charged Nitrogen accepts a hydride from the substrate. In some enzymes the hydride is transferred above (A) and in others below (B) the plane of the pyridine ring. The other proton is usually bound to an amino acid side chain. The reverse is true for the NADH to NAD direction. If you are interested in the details of dehydrogenase enzymes, there are many excellent sections in text books, or reviews: I like the descriptions in Alan Fersht's classic text and this web site will give you more detailed information.NAD, NADP or NAD(P)? The presence of the additional phosphate in NADP presents a significant challenge for an NAD-specific enzyme: such a lot of additional negative charge! In the opposite sense, an NADP-specific enzyme might require a strong set of interactions with the phosphate moiety in order for the enzyme to grasp the substrate (or indeed the transition state), during hydride transfer. In general, enzymes are specific for one or the other cofactor, whilst some like GDH in some species, can handle both with similar effieciency. There is a general rule of thumb however, and that is that anabolic dehydrogenases tend to be NADP-specific, whilst catabolic dehydrogenases prefer NAD. As always, rules of thumb in Biology can be your undoing!
I hope you agree with me that NAD as a suitable candidate as a molecule of the month!
Thanks for all your efforts that you have put in this .very interesting information.i would like to do all the information
ReplyDeleteanaesthesia Machine